In a previous article on this blog have been showed two methods, one geometric and one analitical, for molecule construction in flagstone tessellations.
More in detail, the geometric method can be easily used in a vector graphic software (such as Inkscape) in order to design a flagstone molecule with assigned shape and dimensions.
However, if we want to design a full flagstone tessellation, made up, for instance, of several different flagstone molecules, then the use of the method may became time-consuming,
as a consequence of the increasing number of steps needed to complete each molecule.
Hence the need for a tool able to automatize these steps and to directly provide the final crease pattern of the desired molecule.
That is, the OriFlagTess software.
The software
OriFlagTess is a free software developed in C# under Microsoft Windows environment.
The software "core" is mainly based on the analitical calculation showed in the article mentioned above;
furthermore, analitical translation and rotation formulae are used for the final representation on screen of the molecule.
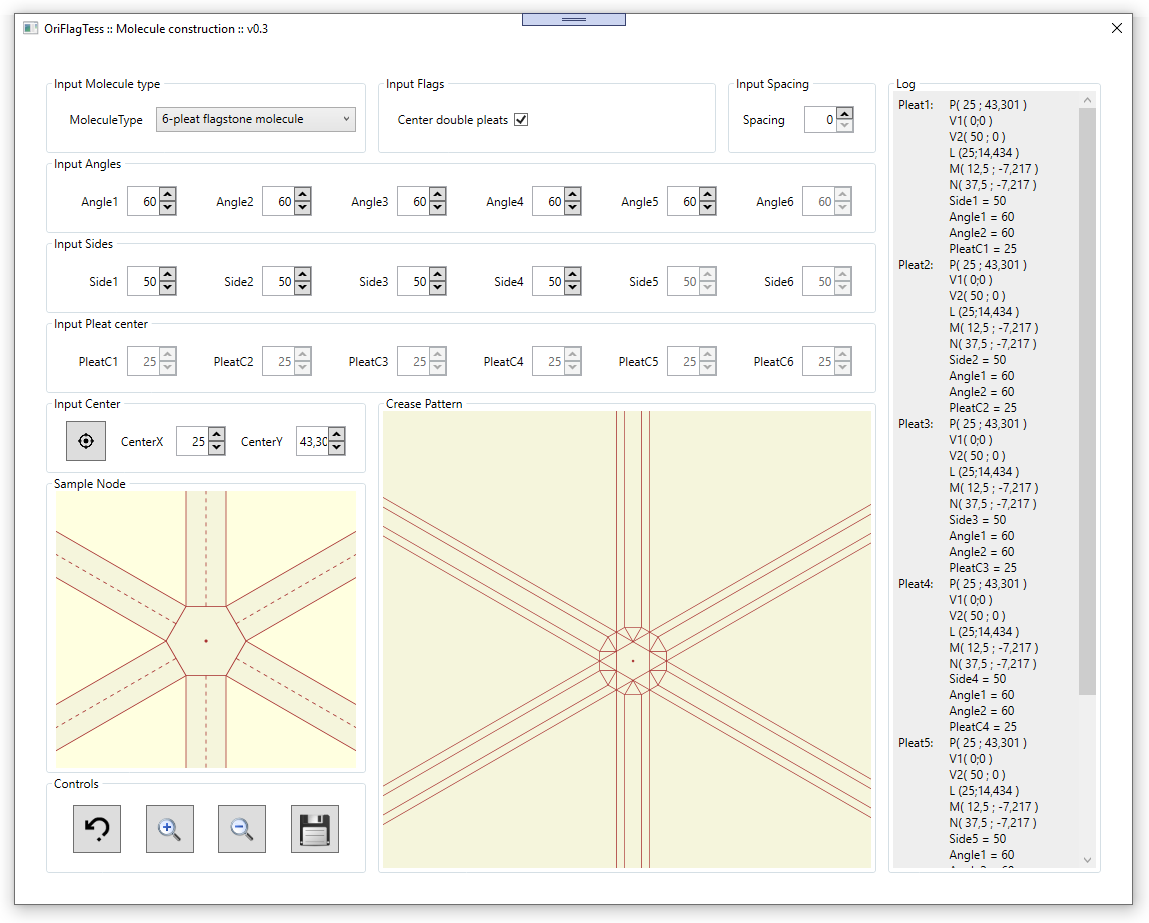
Software consist of a single main window in which is possible to set all the input values, and see a real-time output,
that is the crease pattern of the desired molecule.
Values that is possible to set in input are:
- molecule type
- molecule center coordinates
- molecule sides length (in pixel) 12
- molecule external angles (in degree) 34
- points of each side in which we want to converge the related double pleats (these points are defined as distance from a vertex of each side) ("pleat center")
- the spacing between double pleats in the final model
Every time a value is set or changed, in real-time software do a recalculation and show on screen the new molecule crease pattern.
In the main window we also find a visual box in which is highlighted in real-time the position of the value we are inserting or changing.
Finally, an essential feature is obviously the possibility to save the crease pattern designed in a vectorial format (only SVG at the moment).
Example of a 5-pleat molecule creation
Suppose we want to create a 5-pleat flagstone molecule crease pattern.
We start the software, and, in this order:
- select "5-pleat" as molecule type
- set value of the first $4$ angles, that is $80$, $72$, $80$ and $72$
- set lenght of the first $3$ sides, that is $50$, $60$ and $65$
- set coordinates of the molecule center or click the "auto center" button
Now, in real-time, we obtain the final crease pattern if the molecule.
Then, at this point, the last step is to save it by clicking on the save button.
The following video show the example just described:
Repository and download
The software is freely distributed under GPL v3 license.
It is available on github, at the url https://github.com/UnclePetros/OriFlagTess/.
Here, in the main page, you can find short instructions on software download and install.
If you want to directly download executable zip package, click HERE.
Enjoy!
P.S.: for any feedback on software and its usage you can use the comments section under this post, or the mail address  .
.
Bibliography
- Robert J. Lang, Twists, Tilings, And Tessellations, CRC Press, 2017.
- Bencini, Di Rocco, Geronimo, Il pensiero matematico 1, Ferraro, 1996.
-
To notice the sides set in input, are only the first $n-2$ starting from the bottom and moving anti-clockwise, where $n$ is the number of pleats of the molecule. The other $2$ side are automatically calculated by construction. ↩
-
Values set will match the same pixel number in the final svg image saved at the end of the process. ↩
-
These are the "external" angles of the molecule, that is the angles part of the final node, obtained at the end of the creasing process. ↩
-
Angles set in input are only the first $n-1$ starting from the bottom-left angle and moving anti-clockwise; the last one is automatically given by difference against $360°$ ↩






